Globally, H9N2 viruses have become widespread in wild birds over the last two decades and endemic in poultry in many parts of Eurasia and Africa1, often found co-circulating with high pathogenic avian influenza (HPAI) viruses H5 and H7. In India, the low pathogenic avian influenza (LPAI) H9N2 virus was first reported in 2003 and has been enzootic2-4. The prototype H9N2 virus is an early isolate from turkey flocks in Wisconsin in 1966 [A/turkey/Wisconsin/1/1966(H9N2)]. H9N2 viruses are of low pathogenicity in chickens, which leads to economic loss in poultry-dependant countries due to decreased egg production5. H9N2 viruses can also infect pigs, the mixing vessel for influenza variants.
A retrospective seroprevalence study in India from 1954 to 1981 showed no evidence of H5N1 and H9N2 viruses in birds6. Recent studies have shown transmission linkages of H9N2 viruses in the Middle East, Pakistan, and Bangladesh, with evidence of H9N2 antibodies in poultry workers and emus1,7. Previous studies revealed that the H9N2 viruses from India belonged to the Middle Eastern B sublineage7. Ongoing evolutionary selection pressure and molecular markers associated with mammalian adaptation have been reported in the country4,7. Reassortment between LPAI H9N2 and HPAI H5N1/H7N3 viruses has also been reported in poultries from India7.
Human infection with H9N2 was first reported in 1998 in southern China1,8. To date, globally, there have been 99 cases of human H9N2 virus infections, including two deaths reported to the World Health Organization (WHO) since 20158. Seroprevalence of LPAI H9N2 among the poultry workers has been reported from Pune9. The first laboratory-confirmed human case of H9N2 was reported in a 17-month-old male child from India in 2019. The genomic analysis confirmed that the reassorted H9N2 strain of G1 and H7N3 lineage was susceptible to adamantanes and neuraminidase inhibitors10.
Indian Council of Medical Research (ICMR), in collaboration with the National Influenza Centre (NIC), the apex laboratory at the ICMR- National Institute of Virology (NIV), Pune, established nationwide Pan-India Integrated ARI/SARI surveillance for influenza and SARS-CoV-2 in 2021 through the Virus Research and Diagnostic Laboratories (VRDLs) network across 26 hospital/laboratory sites and 70 community sites11,12.On April 23, 2024, ICMR-National Institute of Cholera and Enteric Diseases (NICED) (currently renamed to ICMR-National Institute for Research in Bacterial Infections) in Kolkata, one of the network laboratories, reported a sample of untypable Influenza A, which was referred to the NIC for confirmation.
Materials & MethodsThe case study was identified as part of the ICMR - Pan India ARI/SARI surveillance through the Virus Research and Diagnostic Laboratories network initiated in 2021 after obtaining ethical clearance from the respective institutes.
Case summaryA three-year-old boy from Malda, West Bengal, who had hyperactive airway disease and was on irregular Metered Dose Inhaler therapy was admitted to Malda Medical College and Hospital (Malda MCH) on February 1, 2024 with severe respiratory distress, recurrent high-grade fever, and abdominal cramps for the last three days. He had a history of recurrent cough and cold since infancy. There was no history of tuberculosis, vomiting, diarrhoea, convulsions, sore throat, rashes over the skin, haematuria, haemoptysis, or ear discharge in the recent past. Investigations at Malda MCH revealed that he was positive for Influenza B and respiratory Adenovirus. He was diagnosed with post-infectious bronchiolitis following viral pneumonia.
The patient was referred to Nil Ratan Sircar Medical College and Hospital (NRS MCH), Kolkata on February 28, 2024 with severe respiratory distress and was put on continuous airway pressure. The patient was shifted to the paediatric intensive care unit (PICU) and was intubated due to Non-Invasive Ventilator failure. The patient was gradually weaned off the ventilator and was put on a high-flow nasal cannula followed by a nasal prong. Investigations at NRS MCH revealed growth of Staphylococcus epidermidis and Candida spp. in blood culture, multifocal consolidation scattered in the bilateral lung in HRCT thorax, and mild widening of cortical sulci and cerebellar follicle cells in brain MRI. The patient was diagnosed with culture-positive sepsis and viral pneumonia with meningo encephalitis. He was finally discharged on May 1, 2024 with bilevel-positive airway pressure support. On inquiry with family members, it was found that the patient had exposure to poultry at home and surroundings. There was no known case of Influenza Like Illness (ILI) in the family or the neighbourhood during the illness of the child.
Laboratory diagnosis at ICMR NICED and ICMR NIV PuneThree consecutive respiratory samples (throat and nasal swabs) were received by ICMR-NICED from NRS MCH on March 5 (VRDL/24/02488), April 18 (VRDL/24/04321), and 25, 2024 (VRDL/24/04645). All three samples were tested for Influenza, SARS-CoV-2, and Respiratory Syncytial Virus (RSV) using in-house NIV multiplex real-time PCR assay11,12. Only the first sample (VRDL/24/02488) was reported as untypable for Influenza A and negative for SARS-CoV-2 and RSV. The remaining two samples, VRDL/24/04321 and VRDL/24/04645 were negative for the respiratory panel. On April 23, 2024, NIC received the Influenza A untypable (VRDL/24/02488) and the second sample (VRDL/24/04321) for confirmation from ICMR-NICED and these samples were tested using in-house NIV multiplex real-time PCR assay11,12 for Influenza, SARS-CoV-2, and RSV. The Influenza-positive sample was further subtyped for seasonal strains (former seasonal influenza A (H1N1), H1N1 pandemic 2009, and H3N2) and non-seasonal avian strains (H5, H7, H9, and H10) as per WHO protocol 202113. Both the samples were also tested for other common respiratory viruses like Human Metapneumovirus, Parainfluenza Virus 1-4, respiratory Adenovirus, Rhinovirus, and enterovirus using the ICMR-NIV duplex assay as described earlier14.
Virus isolationThe influenza-positive respiratory sample was inoculated via the allantoic route in ten-day-old embryonated chicken eggs as per standard WHO protocol15. The inoculated eggs were incubated for 72 h at 37°C in a humidified incubator with daily monitoring for viability, followed by keeping the eggs at 4°C overnight. The allantoic fluids were harvested, and a haemagglutination (HA) assay was carried out for virus detection and titration16,17. Briefly, serial two-fold dilutions of the harvested allantoic fluid were performed with phosphate-buffered saline (PBS) diluent in a V-bottom 96-well plate; 50 µl of 0.5% turkey RBCs (tRBCs) suspension was added in the complete row, and the plate was incubated at room temperature for 30 min. The reciprocal of the last dilution, which showed complete haemagglutination was noted as the HA titre.
50% Egg infectious dose titrationThe infectious titre of the virus isolate was determined in embryonated chicken eggs. Briefly, serial ten-fold dilutions of the virus were inoculated via the allantoic route in four 10-day-old embryonated chicken eggs, followed by incubation at 37°C in a humidified incubator for 72 h. The allantoic fluids were harvested, and the virus was titrated by the HA assay. The EID50 was calculated using the Reed and Muench method with positive haemagglutination as the criteria for positivity15,16.
Haemagglutination inhibition assayThe haemagglutination inhibition (HI) assay was performed to identify the HA subtype15. Briefly, 25 µl of standard H9N2 and H5N1 antiserum, prepared in-house, were serially diluted 2-fold in sterile PBS. A 25 µl of adjusted 8 HAU/50 µl of the isolate was added to all the wells. The plate was incubated at room temperature for 30 min, followed by the addition of 50 µl of tRBCs to all the wells. The plate was then incubated at 4°C for 30 min.
HI assay was also performed to demonstrate the antibody response. The serum samples were obtained from the H9N2 case and family (parents and grandfather) three months post-infection, and HI was performed as per WHO protocol.
Receptor specificity assayThe α-2,3-sialidase assay was carried out to determine the receptor binding preference of the isolate17. Briefly, 50 µl of 10% tRBCs were treated with 1.25 U of the α-2,3-sialidase enzyme (Takara) at 37°C for 1 h. After treatment, the final volume was made up to 0.5% concentration of the RBCs. HA assay was carried out using these treated RBCs.
Fluorescence-based neuraminidase inhibitor assayThe fluorescence-based neuraminidase inhibitor (NAI) assay was performed for the NAI drugs Oseltamivir carboxylate (OC) and Zanamivir susceptibility18. Briefly, the standardised virus dilution was incubated with serial 10-fold dilutions of the drug ranging from 30000 nM to 0.03 nM. The artificial fluorogenic substrate, 2’-(4-methyl-umbelliferyl)-α-D-N-acetylneuraminic acid (MUNANA, Sigma-Aldrich, USA), was added. The generated fluorescence was measured at an excitation wavelength of 365 nm and an emission wavelength of 460 nm (PerkinElmer Victor Nivo Multilabel plate reader). The IC50 values were calculated using the curve-fitting software JASPR (v1.2, Centers for Disease Control and Prevention, USA). The International Society for Influenza and other Respiratory Viruses (ISIRV) reference standard panel A/Mississippi/3/2001 wild type and variant type were used as controls19.
Whole genome sequencing Viral RNA Extraction and rRT-PCRViral RNA was extracted with a magnetic bead-based automated extraction kit. A previously reported universal primer and the rRT-PCR conditions were used to amplify all 8 segments20.
Whole Genome Sequencing using Next-generation sequencingThe whole genome was sequenced using the Illumina MiSeq NGS Platform20. In brief, PCR amplicons of all eight influenza A gene segments were quantified using the Qubit dsDNA BR Assay System (Covaris, Woburn, MA,USA). One ng of the DNA product was processed using the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Adaptor sequences were added to the ends of each fragmented PCR amplicon of approximately 300 bp followed by a 12-cycle PCR to incorporate unique dual indices (i7 and i5) on each DNA fragment, facilitating cluster formation. Subsequent steps included PCR fragment purification and normalisation of the DNA libraries. Equal volumes of each normalised library were pooled to create the barcoded, multiplexed library, which was then sequenced on the Illumina MiSeq system.
Bioinformatics analysisSequencing reads of ⁓150 bp were dynamically trimmed, and FastQC software was used to verify the sequence data21. Iterative refinement meta-assembler (IRMA) pipeline and phylogenetic tree inference were conducted in IQ-TREE version 1.6.9 with 1000 ultra bootstrap support and 1000 SH-like approximate likelihood ratio test (SH-aLRT)22.
Bayesian methodTo calculate the evolutionary rate, a Maximum Clade Credibility (MCC) tree was generated using the Bayesian Markov Chain Monte Carlo approach using the temporal information of the sequence data (BEAST 2.7.6)23,24. The uncorrelated lognormal clock model with the Bayesian Skyline tree prior was used. Three independent chain runs were carried out, each with 100 million generations. 10000 was the sampling frequency for all the runs. Tracer 1.7.1 ( https://beast.community/tracer ) was used to evaluate the chain convergence. FigTree v1.4.4 ( https://tree.bio.ed.ac.uk/software/Figtree/ ) was used to visualize the MCC trees. Further analysis was done with the output files generated by BEAST.
Results Laboratory diagnosisThe clinical sample (VRDL/24/02488) collected on March 5, 2024 was confirmed as Influenza A H9N2 virus subtype with Ct value Inf A: 22; H9: 26 in real-time PCR. The newly identified virus was designated A/India/NIV1519/2024/(H9N2).
Virus isolation and identificationThe allantoic fluid from the inoculated egg showed a 2048 HA titre. The allantoic fluid was harvested and stored at -80°C till further use. The EID50 titre of the isolate was 108.25/200 µl. The isolate was identified as ‘H9’ by the HI assay. The antibodies against the H9N2 virus were detected in the H9N2-positive case with HI titre 1:40, and his parents and grandparents were negative for H9N2-specific antibodies.
Receptor specificity assayNo HA titre was observed in the assay with tRBCs treated with α-2,3-sialidase enzyme. The isolate showed 2048 HA titre with untreated tRBCs. The avian influenza viruses with avian-type receptor specificity do not show haemagglutination with these sialidase-treated RBCs. Thus, the H9N2 isolate showed avian-type receptor specificity (Supplementary Fig. 1).
Fluorescence-based neuraminidase inhibitor assayThe isolate showed IC50 values of 0.05 and 0.28 nM with oseltamivir carboxylate and zanamivir, respectively (Supplementary Fig 2). The reference standards A/Mississippi/3/2001 wild type and variant type showed the IC50 values of 0.29 and 14.97 nM, respectively with oseltamivir, confirming the validity of the assay.
Phylogenetic and molecular analysis of H9N2The subtype was confirmed as H9N2 using the NCBI nucleotide BLAST tool ( https://blast.ncbi.nlm.nih.gov/Blast.cgi ) and GISAID Flusurver analysis. All eight segments showed the highest hits for H9N2 confirming no reassortment gene composition. The HA gene segment of A/India/NIV1519/2024/(H9N2) showed 93.18 per cent similarity with the 2013 Indian isolate A/chicken/India/11FE256/2013(H9N2). Other segments like the nucleoprotein (NP) and NA genes showed high percent similarity with the 2018-19 isolates from Bangladesh A/chicken/Bangladesh/40818/2019(H9N2) and A/chicken/Bangladesh/35278/2018(H9N2), respectively (Supplementary Fig. 3). The gene sequences of all the eight genes of A/India/NIV1519/2024/(H9N2) were submitted to GISAID EPI3452865 to EPI3452872.
HA and NA gene sequences retrieved from the NCBI databases and aligned with the MAFFT alignment tool and the maximum likelihood method (IQTREE) confirmed the virus’s HA gene clusters within the Middle East B lineage of G17 (Fig. 1).
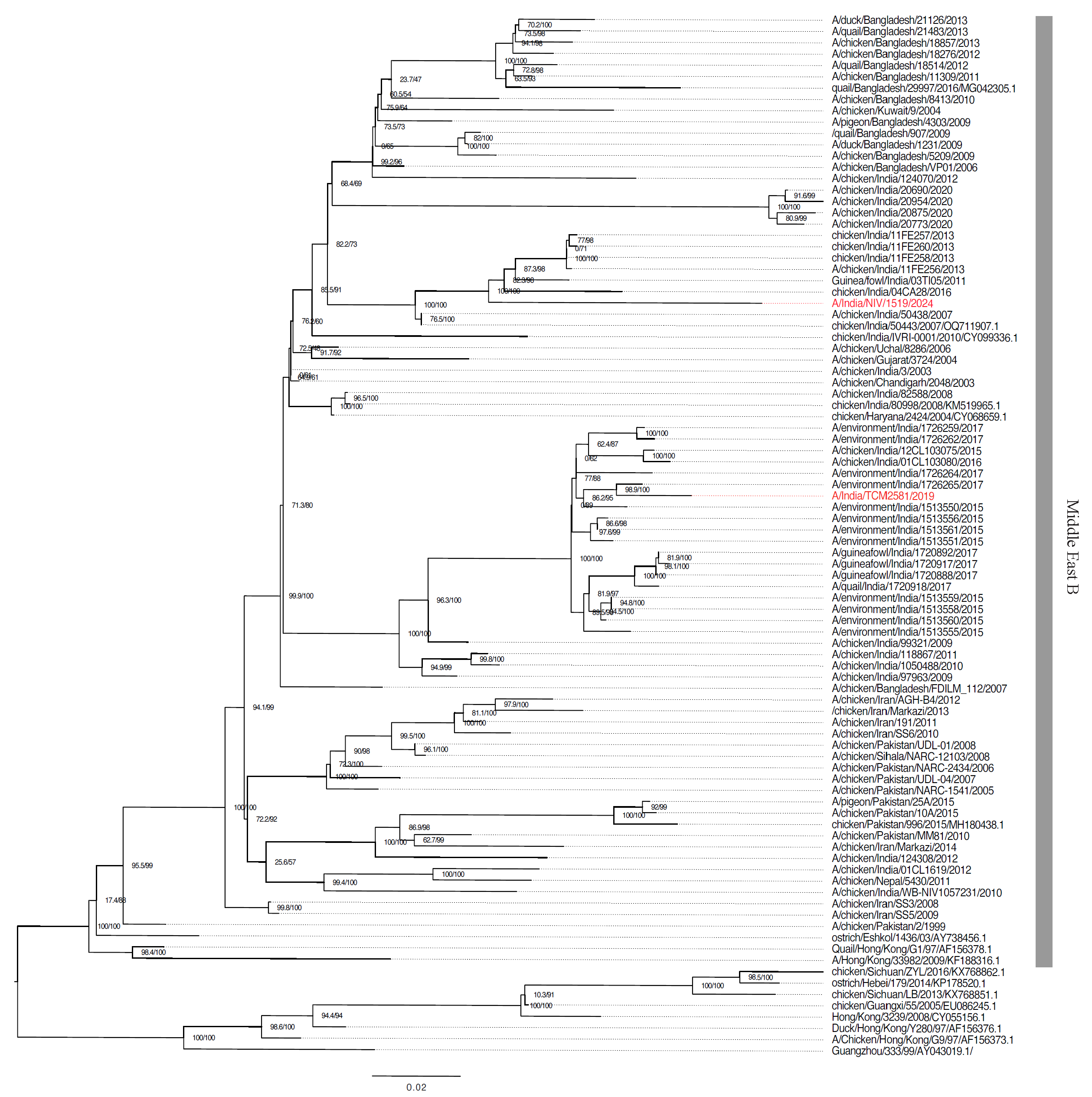
Export to PPT
Significant mutations across various genes with potential impacts on virulence, drug resistance, and host adaptation were catalogued from the GISAID FluSurver module and published inventory of molecular markers (Supplementary Table)25,26. The KSKR/GLF amino acids motif at the cleavage site of HA (335-341 [H9 numbering]) was confirmed in A/India/NIV1519/2024/ (H9N2) strain associated with low pathogenicity in birds. The two most significant mutations A150S in the HA gene and S31N mutation in the MP gene were observed in the study strain. The 150 HA mutation reported previously was associated with increased transmissibility, increased virulence, and host specificity shift. The MP mutation S31N is responsible for the amantadine resistance26,27.
The Indian poultry strains of H9N2 show three clusters of multiple introductions based on HA gene Bayesian evolutionary analyses using BEAST version 2.7.6. The TMRCA of the isolated strain A/India/NIV/1519/2024 with its closest strain was estimated to be in the year 2010 (95% HPD 2008.75-2011.52) (Fig. 2). The evolution rate of Human strain A/India/NIV/1519/2024(H9N2) and the other poultry viruses from India was 4.49 × 10–3 substitutions per site per year.
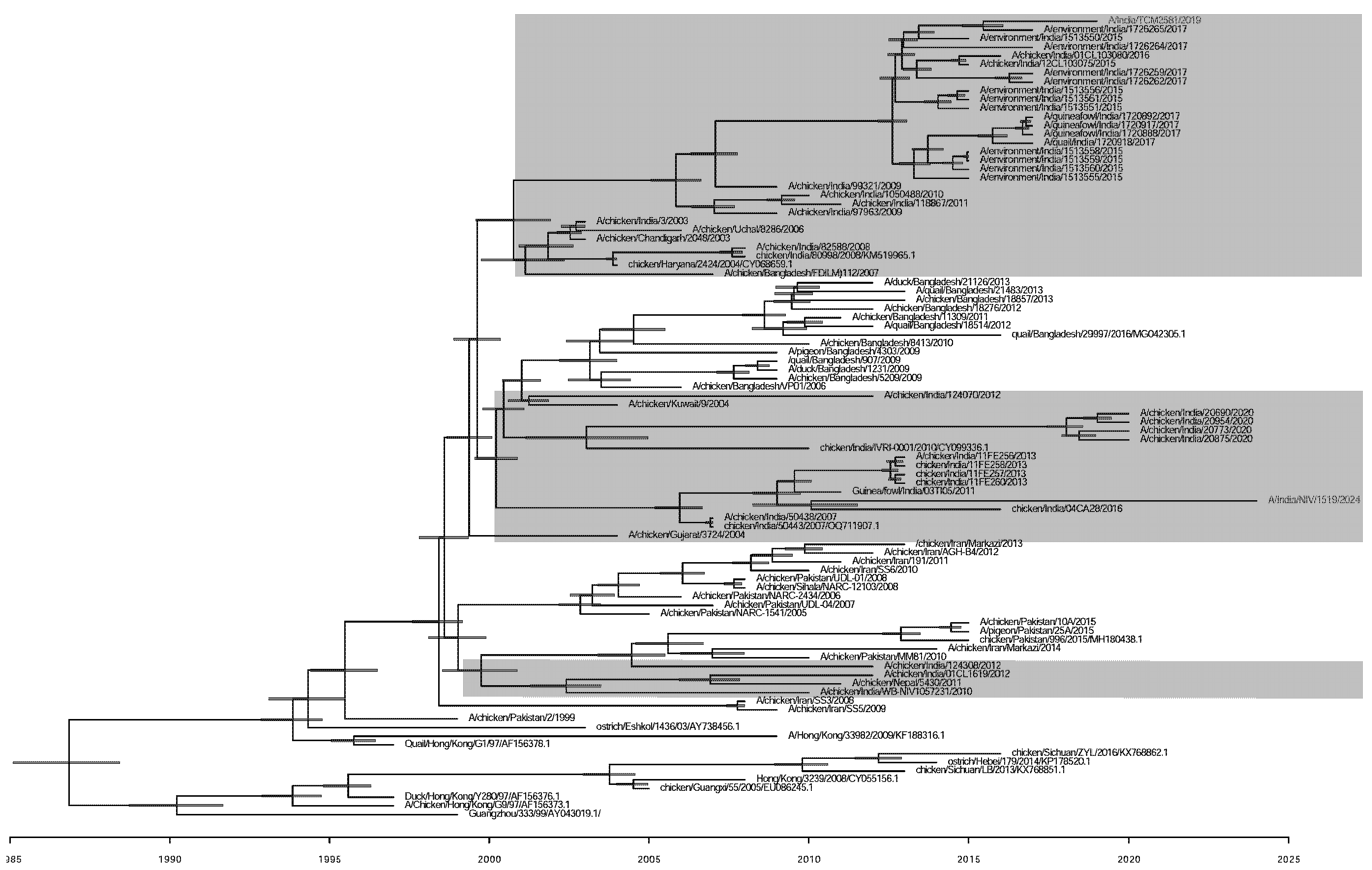
Export to PPT
DiscussionIn the present study, the H9N2 virus was detected and isolated from a human case of respiratory illness, marking the second laboratory-confirmed human infection with the H9N2 in India10. We observed an S31N mutation in the matrix 2 gene of the study strain, known to lead to amantadine resistance in the study strain. We did not observe the known mutations of the NA gene responsible for neuraminidase inhibitors; hence, it remained susceptible to Neuraminidase drugs. The virus remains purely avian-adapted with no markers for mammalian adaptation or human pathogenicity.
No H9 antibodies were found in three close contacts of the infected individual. A joint investigation with the state IDSP team in the local area for ILI/SARI cases yielded no additional confirmed cases. Available epidemiological and virological evidence suggests the virus has not acquired the ability to sustain human-to-human transmission28.
This study’s limitation was that a direct link with poultry exposure could not be established, and other contacts apart from family members could not be investigated. Also, avian influenza surveillance is cost- and labour-intensive, which may limit the ability to get real-time data on circulating strains in traded poultry in India. In addition, establishing epidemiological links for human cases remains challenging.
The ARI/SARI surveillance system established in the country was useful for tracing such unusual events of human infections with H9N2 and H5N1 viruses. Since 2006, over 341 outbreaks of HPAI H5N1 and H5N8 have been reported in avian species, including poultry in India29,30. Reports of H9N2 virus in Indian poultry1-3,6,9,29,30 indicate its presence alongside ongoing H5N1 outbreaks in states like Andhra Pradesh, Maharashtra, Jharkhand, and Kerala31. Gene pool analysis of HA, NA, and PB2 genes from Indian H9N2 viruses (2003–2020) shows they belonged to the Middle Eastern B sublineage of the G1 lineage, alongside viruses from Pakistan, Iran, and Bangladesh7.
Recent years have seen extensive reassortment of H9N2 genes with other viruses like G1, Middle East-B, HPAI H7N3, and HPAI H5N1, creating new genotypes in the Indian subcontinent32. Eastern Asia was the primary source of H9N2 gene segments for the Middle East and Central Asia, with within-country evolution playing a significant role in maintaining viral genetic diversity. This genetic variability is linked to enhanced transmissibility in mammals and increased resistance to antiviral drugs33.
Further, molecular clock and phylogeography analyses of Indian H9N2 viruses reveal transmissions in neighbouring countries, indicating complex transmission patterns. The first reported Indian H910 case had a reassortant genomic composition closely related to Pakistani strains, whereas the current strain did not have such reassortment gene composition. This was based on nucleotide blast results for each gene, which gave the highest hits for H9 viruses. It is also depicted in the phylogeny of all genes of the study strain (Supplementary Fig. 3).
India shares borders with nine countries, many of which have reported H5N1 human cases and numerous H5Nx outbreaks, primarily in poultry34. Neighboring countries have also reported similar subtypes, suggesting H5Nx is endemic in the Asia and Southeast Asia region35,36.
Evolution of the haemagglutinin gene of H5N1 shows multiple clades circulating in the region, with new genotypes resulting from reassortments between LPAI H9N2 and HPAI H5N1 viruses reported in India7. Bangladesh identified a new H5N1 genotype in 2015 with genes from circulating H5N1 and LPAIVs, indicating high reassortment potential37,38. The LPAI H9N2 virus contributes to the genesis of zoonotic AIVs like HPAI H5Nx and emerging human pathogens like H7N939.
Live poultry transportation and mixed trading of domestic animals create environments conducive to gene reassortment, mutation, and interspecies transmission of avian influenza viruses, posing a global concern. Economic and cultural factors leading to close human-poultry interaction underscore the need for targeted, multisectoral, coordinated surveillance efforts for pandemic preparedness using a one-health approach. Identification of a second lab-confirmed case of H9N2 infection in Indians highlights the potential public health threat by avian influenza viruses and the role of the current ARI/SARI surveillance system in tracing such unusual events.
Comments (0)